Lineage first computing: towards a frugal userspace for Linux
. In 1st International Workshop on Low Carbon Computing. .
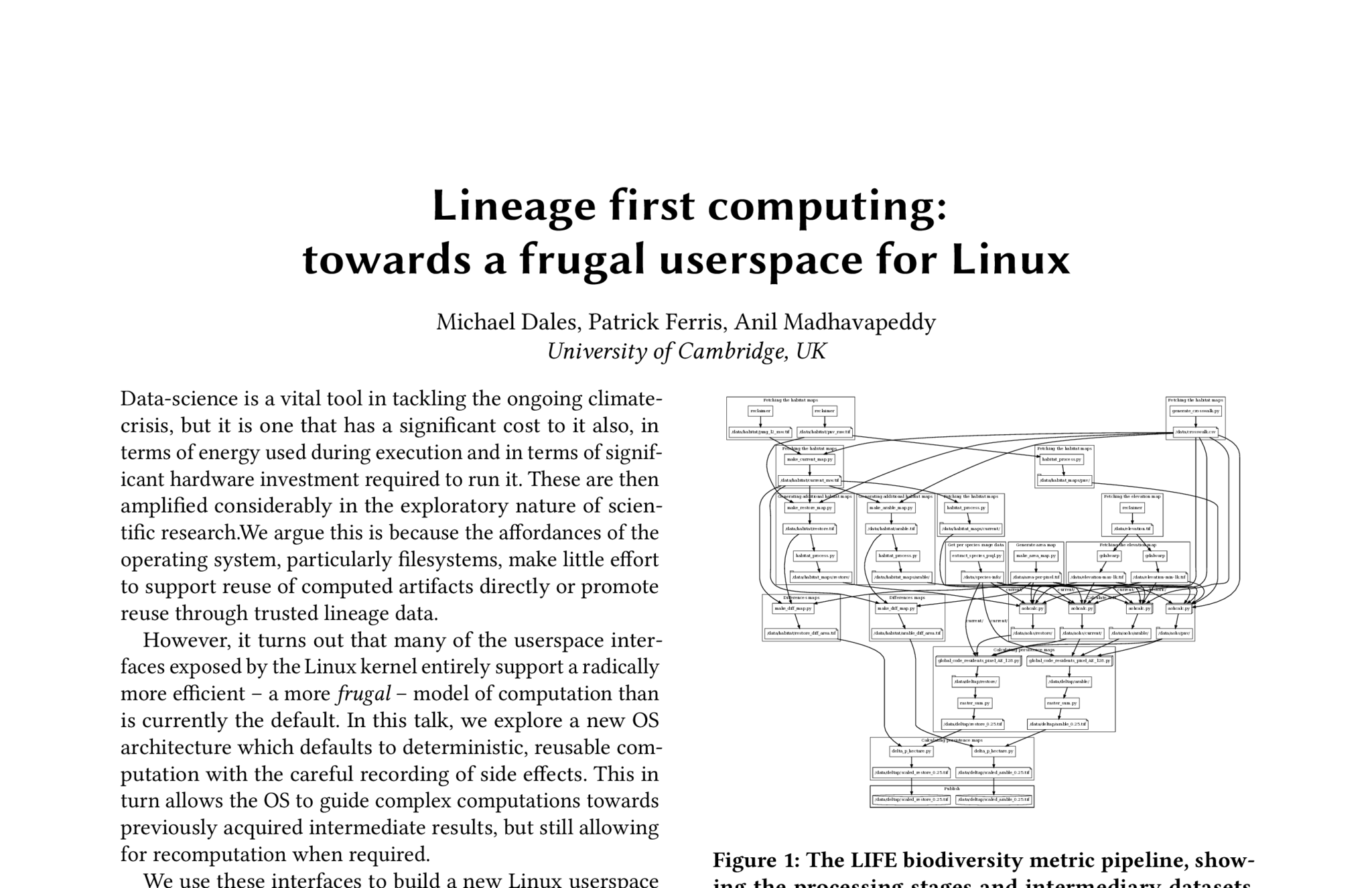
Abstract
Data-science is a vital tool in tackling the ongoing climate-crisis, but it is one that has a significant cost to it also, in terms of energy used during execution and in terms of significant hardware investment required to run it. These are then amplified considerably in the exploratory nature of scientific research. We argue this is because the affordances of the operating system, particularly filesystems, make little effort to support reuse of computed artifacts directly or promote reuse through trusted lineage data.
However, it turns out that many of the userspace interfaces exposed by the Linux kernel entirely support a radically more efficient – a more frugal – model of computation than is currently the default. In this talk, we explore a new OS architecture which defaults to deterministic, reusable computation with the careful recording of side effects. This in turn allows the OS to guide complex computations towards previously acquired intermediate results, but still allowing for recomputation when required.
We use these interfaces to build a new Linux userspace where we put the workflow graph—containing relationships between tools, provenance and labelling—as the core of a system that drives how processes, data, and users interact. Our prototype shell, dubbed Shark, is a data-science first operating environment that is designed to both ensure efficient use of computation and storage resources, and to make it easy for non-experts to create pipeline descriptions from end results post-hoc. It does this by making the lineage graph of a data-pipeline the key concept that ties everything else together: by tracking how the data-pipeline evolves from experimental practice, and tracking what data has already been built and what hasn’t we can prevent re-execution both at development and after publication.